Introduction
In this post, I’ll be implementing K-Means clustering from scratch in Python. This is the seventh post in the “Machine Learning from Scratch” series.
K-Means is one of the most popular unsupervised learning algorithms used for clustering data into groups based on similarity. Unlike the previous algorithms in this series, K-Means doesn’t require labeled data.
K-Means Clustering
K-Means is an unsupervised algorithm that partitions data into K distinct clusters. Each data point belongs to the cluster with the nearest centroid (cluster center).
The algorithm works iteratively:
- Initialize K centroids randomly
- Assign each data point to the nearest centroid
- Update centroids by computing the mean of all points assigned to each cluster
- Repeat steps 2-3 until convergence
The algorithm minimizes the within-cluster sum of squares, making clusters as compact as possible.
Implementation
I’m using numpy for numerical computations and matplotlib for visualization. For testing, I’ll use make_blobs from scikit-learn to generate clustered data.
The KMeans class has the following methods:
__init__: Constructor to set the number of clusters K and maximum iterations.fit: Method to run the K-Means algorithm.predict: Method to assign data points to the nearest cluster.
import numpy as np
from sklearn import datasets
import matplotlib.pyplot as plt
class KMeans:
def __init__(self, K=3, max_iters=100):
self.K = K
self.max_iters = max_iters
self.centroids = []
self.clusters = [[] for _ in range(self.K)]
def fit(self, X):
self.X = X
self.num_samples, self.num_features = X.shape
random_sample_idxs = np.random.choice(self.num_samples, self.K, replace=False)
self.centroids = [self.X[idx] for idx in random_sample_idxs]
for _ in range(self.max_iters):
self.clusters = self._create_clusters(self.centroids)
centroids_old = self.centroids
self.centroids = self._get_centroids(self.clusters)
if self._is_converged(centroids_old, self.centroids):
break
def _create_clusters(self, centroids):
clusters = [[] for _ in range(self.K)]
for idx, sample in enumerate(self.X):
centroid_idx = self._closest_centroid(sample, centroids)
clusters[centroid_idx].append(idx)
return clusters
def _closest_centroid(self, sample, centroids):
distances = [self._euclidean_distance(sample, point) for point in centroids]
closest_index = np.argmin(distances)
return closest_index
def _get_centroids(self, clusters):
centroids = np.zeros((self.K, self.num_features))
for cluster_idx, cluster in enumerate(clusters):
cluster_mean = np.mean(self.X[cluster], axis=0)
centroids[cluster_idx] = cluster_mean
return centroids
def _is_converged(self, centroids_old, centroids):
distances = [
self._euclidean_distance(centroids_old[i], centroids[i])
for i in range(self.K)
]
return sum(distances) == 0
def _euclidean_distance(self, x1, x2):
return np.sqrt(np.sum((x1 - x2) ** 2))
def predict(self, X):
labels = np.zeros(len(X))
for idx, sample in enumerate(X):
labels[idx] = self._closest_centroid(sample, self.centroids)
return labelsNow let’s test the model on synthetic clustered data.
if __name__ == '__main__':
X, _ = datasets.make_blobs(
n_samples=500, n_features=2, centers=3,
cluster_std=1.0, random_state=42
)
model = KMeans(K=3, max_iters=100)
model.fit(X)
labels = model.predict(X)
fig = plt.figure(figsize=(8, 6))
plt.scatter(X[:, 0], X[:, 1], c=labels, cmap='viridis', s=40)
for centroid in model.centroids:
plt.scatter(centroid[0], centroid[1], marker='x',
color='red', s=200, linewidths=3)
plt.show()Let’s visualize the clustering results:
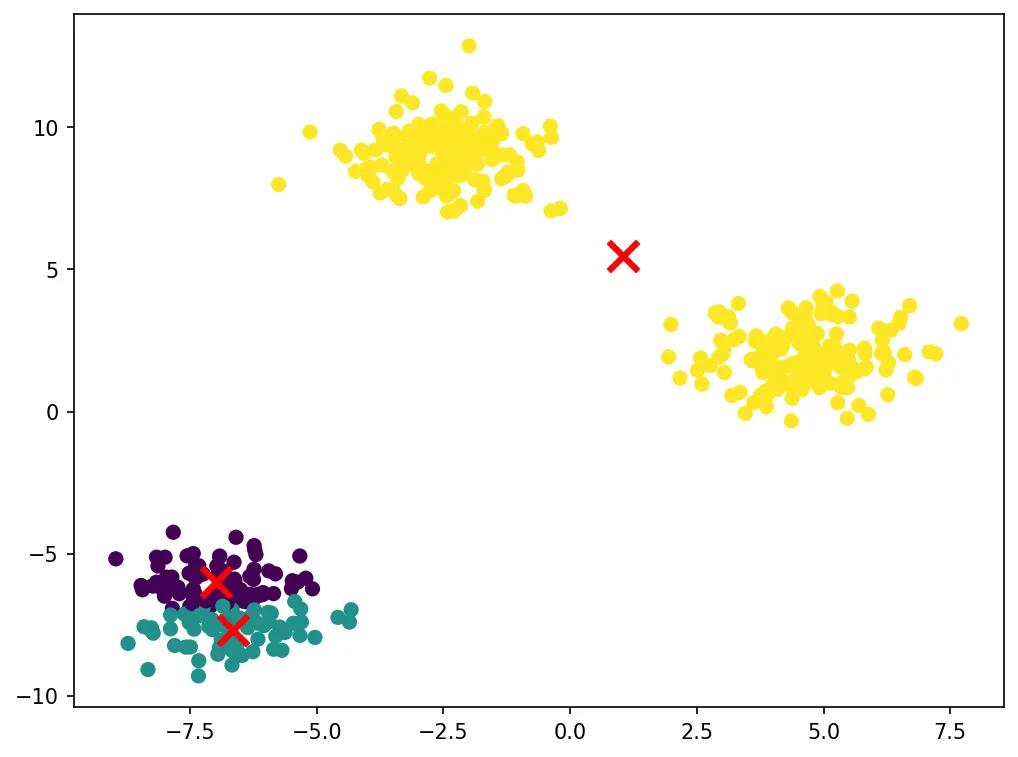
The algorithm successfully identifies the three clusters in the data. The red X marks show the final centroid positions. K-Means works well when clusters are roughly spherical and similar in size.
That’s all for this post. Thanks for reading!